There is an urgent need to improve the reproducibility of (functional) MRI research through transparent reporting, but standardization in this domain is currently lacking. Shared MR images are often only made available after transformation into a derivative data format (e.g. BIDS), and scan session documentation is commonly either manually implemented (e.g. with hand written notes) or neglected entirely.
Scan Session Tool was written to fill this gap, and to be used by neuroscientists, to help them increase transparency and reproducibilty of their MRI research by standardizing scan session documentation and raw data archiving.
Scan Session Tool is a graphical application for documenting (f)MRI scan sessions and automatized data archiving. Information about the scan session itself, used forms and documents, as well as the single measurements can be entered and saved into a protocol file. This information can furthermore be used to copy acquired data (DICOM images as well as optional stimulation protocols and logfiles into a specific hierarchical folder structure for unified archiving purposes, with optional sepcial support for (Turbo-)BrainVoyager.
Installation
Make sure Python 3 (>=3.6, with Tkinter) is installed.
Install pipx:
python3 -m pip install --user pipx
python3 -m pipx ensurepath(On Windows you might need to replace
python3withpy -3).Install Scan Session Tool:
pipx install ScanSessionTool
Usage
Scan Session Tool can be started with the command scansessiontool.
Documentation
Interface
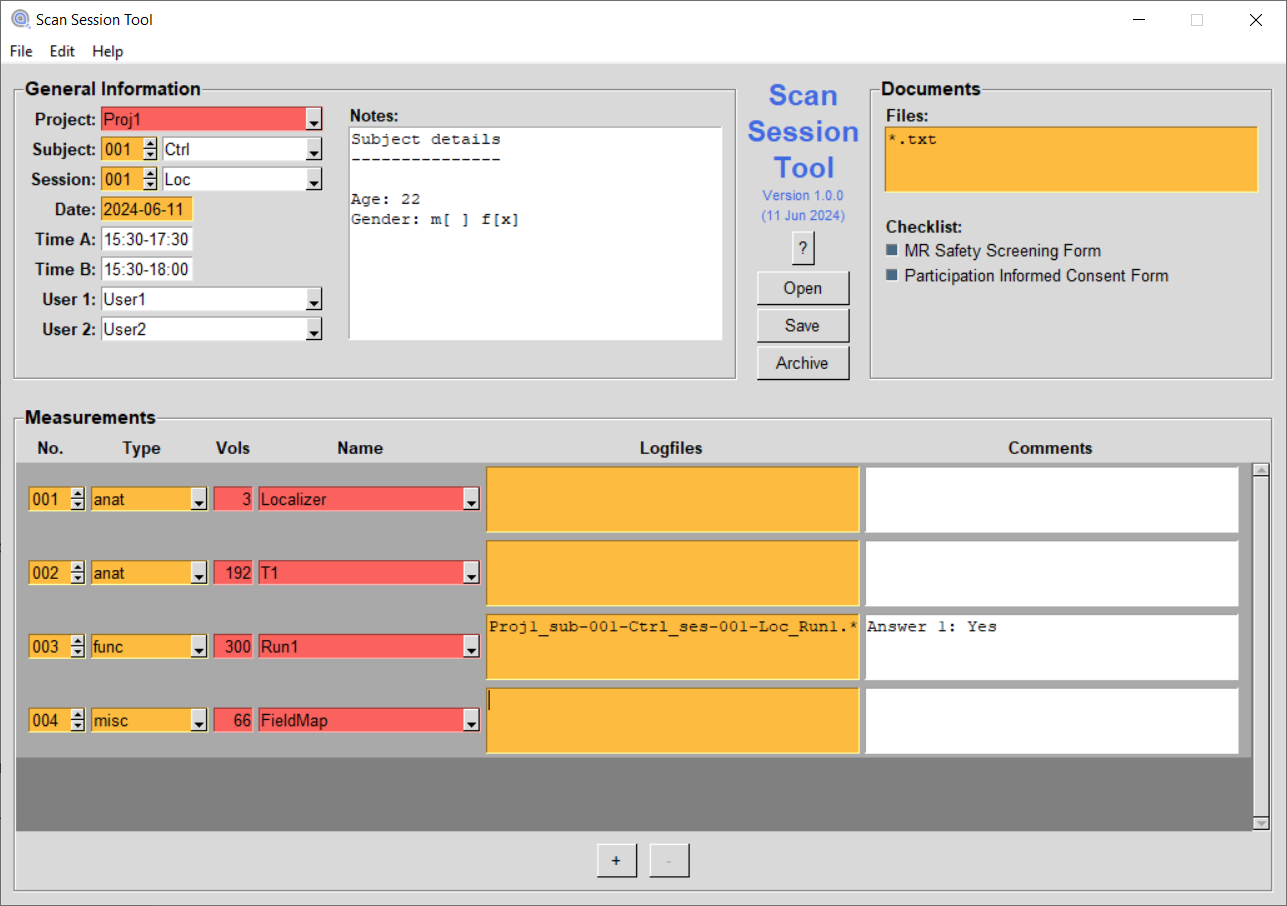
The user interface is organized into three different areas, each holding different information about the scan session, as well as an additional control area for opening and saving session information and for automatically archiving acquired data, based on the session information.
The "General Information" area
This area provides input fields for basic information about the scan session. Some of the fields allow for a selection of pre-specified values taken from a config file (see below), while others take freely typed characters. Fields that are marked with a red background, are mandatory and need to be filled in. Fields that are marked with an orange background are automatically filled in, but need to be checked. The following fields are available:
"Project" - The project identifier
(free-type and selection)
"Subject" - The subject number
(001-999)
- The subject type
(free-type and selection)
"Session" - The session number
(001-999)
- The session type
(free-type and selection)
"Date" - The date of the scan session
(free-type, auto-filled)
"Time A" - The main time period (e.g. official scanner booking)
(free-type)
"Time B" - An additional time period (e.g. actual scanner usage)
(free-type)
"User 1" - The main user (e.g. responsible MR operator)
(free-type and selection)
"User 2" - An additional user (e.g. back-up/buddy)
(free-type and selection)
"Notes" - Any additional notes about the session
(free-type)
The "Documents" area
This area provides checkboxes to specify which forms and documents have been collected from the participant. Additional documents can be specified in a configuration file (see "Config File" section). The following checkboxes are available:
"Files" - A newline separated list of all session logfiles and ad-
ditional documents; wildcard masks (*) will be completed
during archiving
(free-type)
"Checklist" - Checkboxes to specify which forms and documents have been
collected from the participant. Additional documents can
be specified in a configuration file (see "Config File"
section). The following checkboxes are available:
"MR Safety Screening Form" - The (f)MRI scree-
ning form provi-
ded by the scan-
ning institution
"Participation Informed Consent Form" - The official MRI
written consent
form
The "Measurements" area
This area provides several input fields for each measurement of the session. When starting the application, only one (empty) measurement is shown. Clicking on "Add Measurement" will create additional measurements. Fields that are marked with a red background, are mandatory and need to be filled in. Fields that are marked with an orange background are automatically filled in, but need to be checked. The following input fields are available per measurement:
"No" - The number of the measurement
(001-999)
"Type" - "anatomical", "functional" or "misc"
(selection)
"Vols" - The number of volumes of the measurement
(free-type)
"Name" - The name of the measurement
(free-type, selection)
"Logfiles" - A newline separated list of all connected
logfiles; wildcard masks (*) will be completed
during archiving (please note that a stimulation
protocol mask will be included automatically,
based on the session information)
(free-type)
"Comments" - Any additional comments about the measurement
(free-type)
The control area
The control area consists of the following three buttons:
"Open" - Opens previously saved information from a text file
"Save" - Saves the entered session information into a text file
"Archive" - Copies acquired data from a specified source folder into a
target folder at another specified location. Please note that
all data are expected to be within the specified source fol-
der. That is, all DICOM files (*.dcm OR *.IMA; with or with-
out sub-folders), all stimulation protocols and all logfiles.
Optionally, links to the DICOM images in BrainVoyager and
Turbo-BrainVoyager formats can be created. Turbo-BrainVoyager
files and data will be manipulated to work in the target
directory.
The data will be copied into the following folder hierarchy:
DICOMs -->
<Project>/sub-<Subject>/ses-<Session>/<Type>/
<No>-<Name>/<DICOM>/
Logfiles -->
<Project>/sub-<Subject>/ses-<Session>/<Type>/
<No>-<Name>/
Files -->
<Project>/sub-<Subject>/ses-<Session>/
BrainVoyager files (links only, optional) -->
<Project>/sub-<Subject>/ses-<Session>/<BV>/
Turbo-BrainVoyager files (links only, optional) -->
<Project>/sub-<Subject>/ses-<Session>/<TBV>/
Scan Session Protocol -->
<Project>/sub-<Subject>/ses-<Session>/
Config File
A configuration file can be created to pre-define the values to be used as selection options for the "Group", "Session", "Certified User", "Backup Person", "Notes", the measurement "Name", "Vols" and "Comments" on a per project basis, as well as additional items in the "Documents" section. The Scan Session Tool will look for a configuration file with the name "sst.yaml", located in the current working directory or in the $HOME folder.
The syntax is YAML. Here is an example:
Project 1:
SubjectTypes:
- Group1
- Group2
SessionTypes:
- Sess1
- Sess2
Users:
- User1
- User2
Backups:
- User1
- User2
Notes: |
Subject details
---------------
Age:
Gender: m[ ] f[ ]
Files:
- "*.txt"
Checklist:
- Pre-Scan Questionnaire
- Post-Scan Questionnaire
Measurements anatomical:
- Name: Localizer
Vols: 3
- Name: Anatomy
Vols: 192
Measurements functional:
- Name: Run1
Vols: 300
Comments: |
Answer 1:
- Name: Run2
Vols: 400
Comments: |
Answer 2:
- Name: Run3
Vols: 200
Comments: |
Answer 3:
Measurements misc:
- Name: Run1incomplete
Vols:
Comments:
Project 2:
SubjectTypes:
- GroupA
- GroupB
SessionTypes:
- SessA
- SessB
Users:
- UserA
- UserB
Backups:
- UserA
- UserB
Notes: |
Subject details
---------------
Age:
Gender: m[ ] f[ ]
Files:
- "*.txt"
Checklist:
- Participation Reimbursement Form
Measurements anatomical:
- Name: Localizer
Vols: 3
- Name: MPRAGE
Vols: 192
Measurements functional:
- Name: RunA
Vols: 300
Comments: |
Answer 1:
- Name: RunB
Vols: 400
Comments: |
Answer 2:
- Name: RunC
Vols: 200
Comments: |
Answer 3:
Measurements misc:
- Name: RunAImcomplete
Vols:
Comments:
Tutorial
For a detailed step-by-step guide on how to use the Scan Session Tool you can follow the link: Scan Session Tool tutorial .